
If you already heard about this package, good for you. When we have lots of data points to plot, this can run into the issue of. Create a scatter plot with varying marker point size and color. (A 400圆00 image only has 240K dots.) So the easiest thing to do would be to take a sample of say, 1000 points, from your data: import random deltasamplerandom.sample(delta,1000) and just plot that. Implementing and sharing this amazingly fast package! A scatter plot (aka scatter chart, scatter graph) uses dots to represent values. Unless your graphic is huge, many of those 3 million points are going to overlap. Would highly recommend using scattermore. So, if you have to plot a huge amount of points into a scatterplot, as I often do, I The overall speed up now is of ~13x: from 13.55 s to ~1 s! require ( scattermore ) system.time ( print ( ggplot ( pdata, aes ( x = x, y = y )) + geom_scattermore ())) user system elapsed 0.987 0.060 1.047 Time of writing this post), which uses a C script to rasterize the dots as a bitmap and 1 I have an array of 500000 samples i.e., the data's shape is (500000, 3) where the first two columns represent x-coordinate and y- coordinate, and the third column is Label values to which the datapoint (X,Y) belongs.

His new R package scattermore (last commit to the package was on Jan 31st, 2021, at the Then, I found another StackOverflow answer, with a user recommending In contrast to line graphs, each point is independent. This provides a ~5x speed up, from 13.6 s to less than 3 s! scattermore is faster Scatter plots are used to graph data along two continuous dimensions. Actually Ive 3 vectors with the same dimension and I use to plot in the following way. system.time ( print ( ggplot ( pdata, aes ( x = x, y = y )) + geom_point ( pch = '.' ))) user system elapsed 2.688 0.100 2.787 Scatter plot with a huge amount of data Ask Question Asked 12 years, 8 months ago Modified 12 years, 6 months ago Viewed 55k times 24 I would like to use Matplotlib to generate a scatter plot with a huge amount of data (about 3 million points). Recommending to use the pch='.' option to plot data points as non-aliased single One of the tips I found on the web comes from a StackOverflow answer, How long would the default R plot() and ggplot methods take to plot this? system.time ( with ( pdata, plot ( x, y ))) user system elapsed 11.481 0.048 11.530 require ( ggplot2 ) system.time ( print ( ggplot ( pdata, aes ( x = x, y = y )) + geom_point ())) user system elapsed 13.331 0.220 13.552Īnd here is our starting point: R would take around 11.5 s and ggplot even longer, Let’s start by generating a dataset of 1 million X and Y coordinates, normallyĭistributed: require ( data.table ) pdata = data.table ( x = rnorm ( 1e6 ), y = rnorm ( 1e6 )) Today, I finally got tired of it and went down a rabbit hole of DDG searches (yes, I useĭuckDuckGo, and you should too!). More than 5 min for it to be generated and exported to a png file. Only then I would generate the final plot using all data points, sometimes waiting
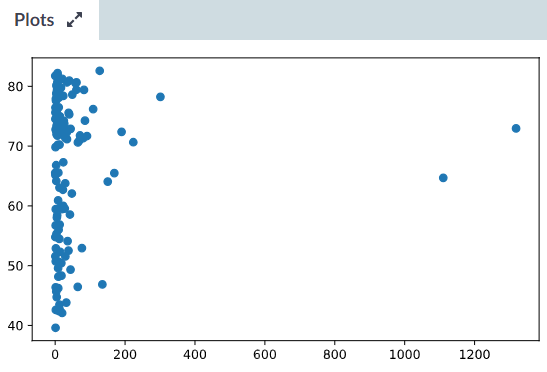
Until now, I usually plotted just a few randomly selected points while fixing the figure Quite the bottleneck: plotting can take forever. But, when handling such large amounts of data, I always encounter My tool of choice for plotting is always R, and more specifically the grammar of graphs One of the easiest and simplest ways to make your graphs stand out is to change the default background.
#PYTHON SCATTER PLOT WITH THOUSANDS OF POINTS CODE#
If you’re like me and you often forget the precise code to format plots, this piece is written specifically for you. A scatter plot is a visual representation of how two variables relate to each other. Have you ever had to generate a scatterplot with one million points, or more? As aīioinformatician working in the academia, and specifically on large datasets, this Matplotlib is the most extensive plotting library in python, arguably one of the most frequently used. title ( 'Overplotting? Show putative structure', loc = 'left' ) plt. legend ( loc = 'lower right', markerscale = 2 ) # titles plt.


 0 kommentar(er)
0 kommentar(er)
